Page 5 - Krész, Miklós, and Andrej Brodnik (eds.). MATCOS-13. Proceedings of the 2013 Mini-Conference on Applied Theoretical Computer Science. Koper: University of Primorska Press, 2016.
P. 5
ficient implementation of algorithms for solving
subgraph isomorphism problem in cheminformatics
Mónika Vigula
Eötvös Loránd University
Budapest, Hungary
vigula.monika@gmail.com
ABSTRACT (mostly hundreds of thousands or millions) and a query
molecule are given. We have to find all the target molecules
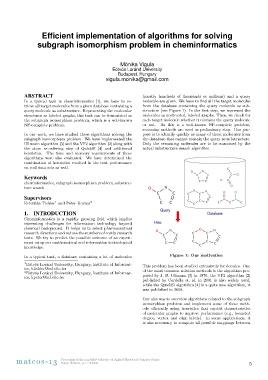
In a typical task in cheminformatics [1], we have to re- from the database containing the query molecule as sub-
trieve all target molecules from a given database containing a structure (see Figure 1). In the first step, we represent the
query molecule as substructure. Representing the molecular molecules as undirected, labeled graphs. Then, we check for
structures as labeled graphs, this task can be formulated as each target molecule whether it contains the query molecule
the subgraph isomorphism problem, which is a well-known or not. As this is a well-known NP-complete problem,
NP-complete problem. screening methods are used as preliminary step. Our pur-
pose is to identify quickly as many of those molecules from
In our work, we have studied three algorithms solving the the database that cannot contain the query as substructure.
subgraph isomorphism problem. We have implemented the Only the remaining molecules are to be examined by the
Ullmann algorithm [5] and the VF2 algorithm [2] along with actual substructure search algorithm.
the atom re-ordering step of QuickSI [4] and additional
heuristics. The time and memory requirements of these Figure 1: Our motivation
algorithms were also evaluated. We have determined the
combination of heuristics resulted in the best performance This problem has been studied extensively for decades. One
on real data sets as well. of the most common solution methods is the algorithm pro-
posed by J. R. Ullmann [5] in 1976, the VF2 algorithm [2]
Keywords published by Cordella et. al. in 2001 is also widely used,
while the QuickSI algorithm [4] is a quite new algorithm, it
cheminformatics, subgraph isomorphism problem, substruc- was published in 2008.
ture search Our aim was to overview algorithms related to the subgraph
isomorphism problem and implement some of these meth-
Supervisors ods efficiently using heuristics that exploit characteristics
of molecular graphs to improve performance (e.g., bounded
Krisztia´n Tichler1 and P´eter Kova´cs2 degree, vertex and edge labels). In some applications, it
is also necessary to compute all possible mappings between
1. INTRODUCTION
Cheminformatics is a rapidly growing field which implies
interesting challenges for information technology beyond
chemical background. It helps us to select pharmaceutical
research directions and reduce the numbers of costly research
tests. We try to predict the possible outcome of an experi-
ment using our mathematical and information technological
knowledge.
In a typical task, a database containing a lot of molecules
1E¨otv¨os Lora´nd University, Hungary, Institute of Informat-
ics, ktichler@inf.elte.hu
2Eo¨tv¨os Lora´nd University, Hungary, Institute of Informat-
ics, kpeter@inf.elte.hu
m a t c o s -1 3 Proceedings of the 2013 Mini-Conference on Applied Theoretical Computer Science 5
Koper, Slovenia, 10-11 October
subgraph isomorphism problem in cheminformatics
Mónika Vigula
Eötvös Loránd University
Budapest, Hungary
vigula.monika@gmail.com
ABSTRACT (mostly hundreds of thousands or millions) and a query
molecule are given. We have to find all the target molecules
In a typical task in cheminformatics [1], we have to re- from the database containing the query molecule as sub-
trieve all target molecules from a given database containing a structure (see Figure 1). In the first step, we represent the
query molecule as substructure. Representing the molecular molecules as undirected, labeled graphs. Then, we check for
structures as labeled graphs, this task can be formulated as each target molecule whether it contains the query molecule
the subgraph isomorphism problem, which is a well-known or not. As this is a well-known NP-complete problem,
NP-complete problem. screening methods are used as preliminary step. Our pur-
pose is to identify quickly as many of those molecules from
In our work, we have studied three algorithms solving the the database that cannot contain the query as substructure.
subgraph isomorphism problem. We have implemented the Only the remaining molecules are to be examined by the
Ullmann algorithm [5] and the VF2 algorithm [2] along with actual substructure search algorithm.
the atom re-ordering step of QuickSI [4] and additional
heuristics. The time and memory requirements of these Figure 1: Our motivation
algorithms were also evaluated. We have determined the
combination of heuristics resulted in the best performance This problem has been studied extensively for decades. One
on real data sets as well. of the most common solution methods is the algorithm pro-
posed by J. R. Ullmann [5] in 1976, the VF2 algorithm [2]
Keywords published by Cordella et. al. in 2001 is also widely used,
while the QuickSI algorithm [4] is a quite new algorithm, it
cheminformatics, subgraph isomorphism problem, substruc- was published in 2008.
ture search Our aim was to overview algorithms related to the subgraph
isomorphism problem and implement some of these meth-
Supervisors ods efficiently using heuristics that exploit characteristics
of molecular graphs to improve performance (e.g., bounded
Krisztia´n Tichler1 and P´eter Kova´cs2 degree, vertex and edge labels). In some applications, it
is also necessary to compute all possible mappings between
1. INTRODUCTION
Cheminformatics is a rapidly growing field which implies
interesting challenges for information technology beyond
chemical background. It helps us to select pharmaceutical
research directions and reduce the numbers of costly research
tests. We try to predict the possible outcome of an experi-
ment using our mathematical and information technological
knowledge.
In a typical task, a database containing a lot of molecules
1E¨otv¨os Lora´nd University, Hungary, Institute of Informat-
ics, ktichler@inf.elte.hu
2Eo¨tv¨os Lora´nd University, Hungary, Institute of Informat-
ics, kpeter@inf.elte.hu
m a t c o s -1 3 Proceedings of the 2013 Mini-Conference on Applied Theoretical Computer Science 5
Koper, Slovenia, 10-11 October